Schwartz, E., Fritsch, J. & Friedrich, B. H2-metabolizing prokaryotes (Springer Berlin Heidelberg, 2013).
Greening, C. et al. Genome and metagenome surveys of hydrogenase diversity indicate H2 is a widely-utilised energy source for microbial growth and survival. Isme J. 10, 761–777 (2016).
Cook, G. M., Greening, C., Hards, K. & Berney, M. In Advances in Bacterial Pathogen Biology (ed. Poole, R. K. ) 65, 1–62 (Academic Press, 2014).
Lane, N., Allen, J. F. & Martin, W. How did LUCA make a living? Chemiosmosis in the origin of life. BioEssays 32, 271–280 (2010).
Weiss, M. C. et al. The physiology and habitat of the last universal common ancestor. Nat. Microbiol. 1, 16116 (2016).
Lubitz, W., Ogata, H., Rüdiger, O. & Reijerse, E. Hydrogenases. Chem. Rev. 114, 4081–4148 (2014).
Peters, J. W. et al. [FeFe]- and [NiFe]-hydrogenase diversity, mechanism and maturation. Biochim. Biophys. Acta – Mol. Cell Res. 1853, 1350–1369 (2014).
Carbonero, F., Benefiel, A. C. & Gaskins, H. R. Contributions of the microbial hydrogen economy to colonic homeostasis. Nat Rev Gastroenterol Hepatol 9, 504–518 (2012).
Greening, C. et al. Atmospheric hydrogen scavenging: from enzymes to ecosystems. Appl. Environ. Microbiol. 81, 1190–1199 (2015).
Levin, D. B., Pitt, L. & Love, M. Biohydrogen production: prospects and limitations to practical application. Int. J. Hydrogen Energy 29, 173–185 (2004).
Cracknell, J. A., Vincent, K. A. & Armstrong, F. A. Enzymes as working or inspirational catalysts for fuel cells and electrolysis. Chem. Rev. 108, 2439–2461 (2008).
Shima, S. et al. The crystal structure of [Fe]-Hydrogenase reveals the geometry of the active site. Science 321, 572–575 (2008).
Lenz, O. & Friedrich, B. A novel multicomponent regulatory system mediates H2 sensing in Alcaligenes eutrophus. Proc. Natl. Acad. Sci. USA 95, 12474–12479 (1998).
Greening, C., Berney, M., Hards, K., Cook, G. M. & Conrad, R. A soil actinobacterium scavenges atmospheric H2 using two membrane-associated, oxygen-dependent [NiFe] hydrogenases. Proc. Natl. Acad. Sci. USA 111, 4257–4261 (2014).
Schuchmann, K. & Müller, V. A bacterial electron-bifurcating hydrogenase. J. Biol. Chem. 287, 31165–31171 (2012).
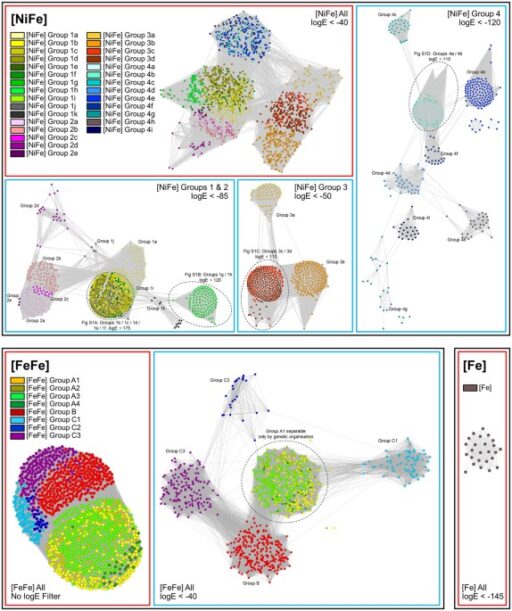
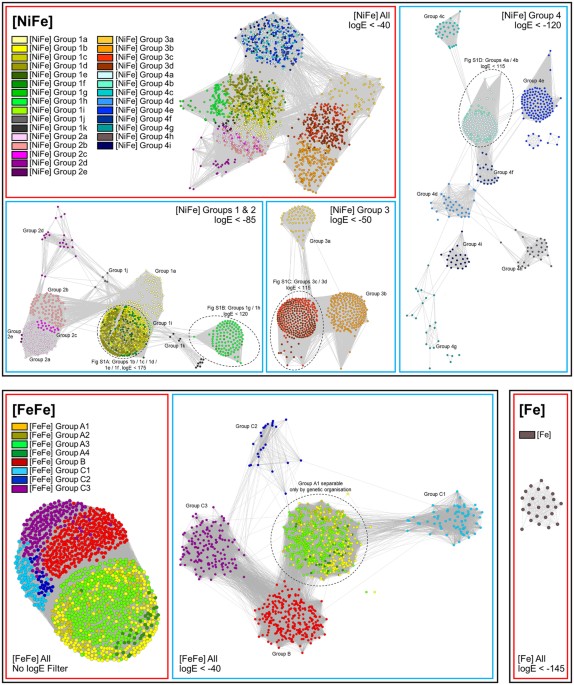
Vignais, P. M., Billoud, B. & Meyer, J. Classification and phylogeny of hydrogenases. Fems Microbiol. Rev. 25, 455–501 (2001).
Calusinska, M., Happe, T., Joris, B. & Wilmotte, A. The surprising diversity of clostridial hydrogenases: a comparative genomic perspective. Microbiology 156, 1575–1588 (2010).
Atkinson, H. J., Morris, J. H., Ferrin, T. E. & Babbitt, P. C. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. Plos One 4, e4345 (2009).
Ahmed, F. H. et al. Sequence-structure-function classification of a catalytically diverse oxidoreductase superfamily in mycobacteria. J. Mol. Biol. 427, 3554–3571 (2015).
Ney, B. et al. The methanogenic redox cofactor F420 is widely synthesized by aerobic soil bacteria. Isme J., 10.1038/ismej.2016.100 (2016).
Stetter, K. O. Archaeoglobus fulgidus gen. nov., sp. nov.: a new taxon of extremely thermophilic archaebacteria. Syst. Appl. Microbiol. 10, 172–173 (1988).
Deppenmeier, U. & Blaut, M. Analysis of the vhoGAC and vhtGAC operons from Methanosarcina mazei strain Gö1, both encoding a membrane-bound hydrogenase and a cytochrome b. Eur. J. Biochem. 269, 261–269 (1995).
Kim, Y. J. et al. Formate-driven growth coupled with H2 production. Nature 467, 352–355 (2010).
McTernan, P. M. et al. Intact functional fourteen-subunit respiratory membrane-bound [NiFe]-hydrogenase complex of the hyperthermophilic archaeon Pyrococcus furiosus. J. Biol. Chem. 289, 19364–19372 (2014).
Lie, T. J. et al. Essential anaplerotic role for the energy-converting hydrogenase Eha in hydrogenotrophic methanogenesis. Proc. Natl. Acad. Sci. USA 109, 15473–15478 (2012).
Auernik, K. S. & Kelly, R. M. Physiological versatility of the extremely thermoacidophilic archaeon Metallosphaera sedula supported by transcriptomic analysis of heterotrophic, autotrophic and mixotrophic growth. Appl. Environ. Microbiol. 76, 931–935 (2010).
Giaveno, M. A., Urbieta, M. S., Ulloa, J. R., González Toril, E. & Donati, E. R. Physiologic versatility and growth flexibility as the Main characteristics of a novel thermoacidophilic Acidianus strain isolated from Copahue geothermal area in Argentina. Microb. Ecol. 65, 336–346 (2012).
Poudel, S. et al. Unification of [FeFe]-hydrogenases into three structural and functional groups. Biochim. Biophys. Acta (BBA)-General Subj., 10.1016/j.bbagen.2016.05.034 (2016).
Marchler-Bauer, A. & Bryant, S. H. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32, W327–W331 (2004).
Berney, M., Greening, C., Hards, K., Collins, D. & Cook, G. M. Three different [NiFe] hydrogenases confer metabolic flexibility in the obligate aerobe Mycobacterium smegmatis. Environ. Microbiol. 16, 318–330 (2014).
Greening, C. et al. Persistence of the dominant soil phylum Acidobacteria by trace gas scavenging. Proc. Natl. Acad. Sci. 112, 10497–10502 (2015).
Chen, Z. et al. Phaeodactylibacter xiamenensis gen. nov., sp. nov., a member of the family Saprospiraceae isolated from the marine alga Phaeodactylum tricornutum. Int. J. Syst. Evol. Microbiol. 64, 3496–3502 (2014).
Koch, H. et al. Growth of nitrite-oxidizing bacteria by aerobic hydrogen oxidation. Science 345, 1052–1054 (2014).
Carere, C. R. et al. Growth and persistence of methanotrophic bacteria by aerobic hydrogen respiration. Proc. Natl. Acad. Sci. USA (2016).
Haroon, M. F. et al. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500, 567–570 (2013).
Evans, P. N. et al. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science 350, 434–438 (2015).
Brown, C. T. et al. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 523, 208–211 (2015).
Spang, A. et al. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521, 173–179 (2015).
Eloe-Fadrosh, E. A. et al. Global metagenomic survey reveals a new bacterial candidate phylum in geothermal springs. Nat Commun 7 (2016).
Wilson, M. C. et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 506, 58–62 (2014).
Greening, C. & Cook, G. M. Integration of hydrogenase expression and hydrogen sensing in bacterial cell physiology. Curr. Opin. Microbiol. 18, 30–38 (2014).
Greening, C. et al. Physiology, biochemistry and applications of F420- and Fo-dependent redox reactions. Microbiol. Mol. Biol. Rev. 80, 451–493 (2016).
Markowitz, V. M. et al. IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Research 40, D115–D122 (2012).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Cover, T. & Hart, P. Nearest neighbor pattern classification. Ieee Trans. Inf. Theory 13 (1967).
Constant, P., Chowdhury, S. P., Pratscher, J. & Conrad, R. Streptomycetes contributing to atmospheric molecular hydrogen soil uptake are widespread and encode a putative high-affinity [NiFe]-hydrogenase. Environ. Microbiol. 12, 821–829 (2010).
Hamann, E. et al. Environmental Breviatea harbour mutualistic Arcobacter epibionts. Nature 534, 254–258 (2016).
Sousa, F. L., Neukirchen, S., Allen, J. F., Lane, N. & Martin, W. F. Lokiarchaeon is hydrogen dependent. Nat. Microbiol. 1, 16034 (2016).