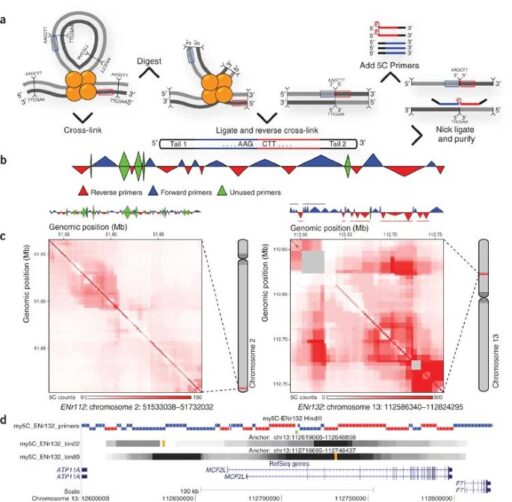
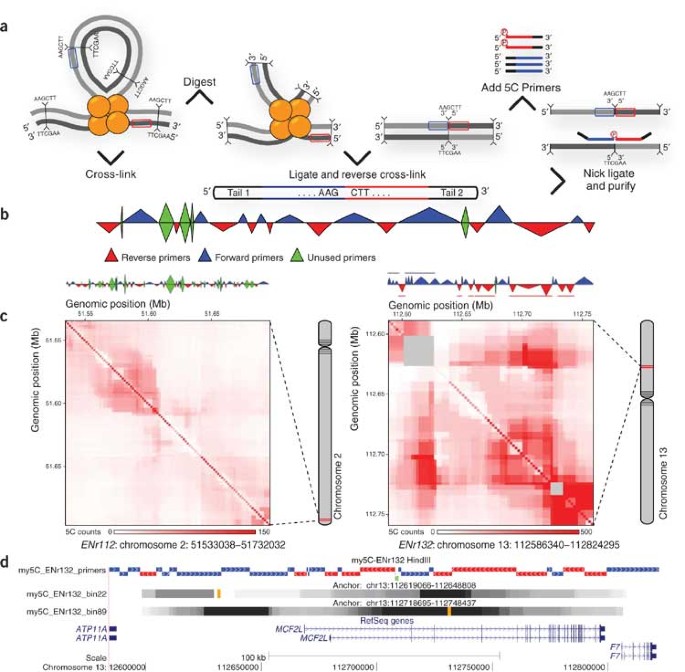
To the Editor: The three-dimensional arrangement of chromosomes is critical for genome regulation. Chromosome organization can be studied using chromosome conformation capture (3C)-based assays1,2. The ‘3C carbon copy’ (5C) approach is an adaptation of 3C for high-throughput analysis of interaction networks and three-dimensional folding of chromosomes3.
My5C comprises two modules. The my5C.primers module is used to design 5C primers for restriction fragments throughout user-defined genomic regions (Supplementary Data 5). For analysis of overall three-dimensional conformation, alternating primer design schemes3 (Fig. 1b) can be selected (Supplementary Data 6). For studies of networks of interactions between specific genomic elements–for example, promoters and enhancers3–users can upload files containing genomic coordinates of these elements and my5C.primers will design forward and reverse primers for the two sets (Supplementary Data 7). Primer designs can be downloaded along with a custom microarray probe set for detection of all interrogated interactions.